Circuits in the import graph
The first thing to note is that usually we'll be talking about a directed graph, but there are certain cases where a Python package doesn't have to adhere to these. One prominent example is forward references (a problem I've encountered when thoroughly type annotating libraries).
See the
ForwardReftype, this discussion on SO, the section on them in PEP 484, and PEP 563.
Modules importing each other in Python is known as 'circular' imports (i.e. circuits or cycles in
graph theory terminology).
As described here
the trick I used was to use an if TYPE_CHECKING: statement. It gets ugly though,
so let's not dwell any further on it.
A prototype
To demonstrate the import_deps library, I put a single-arg CLI as an entrypoint to a package
I generated from py-pkg-cc-template (a cookiecutter package template whose setup I described in
the Package templating series).
Its key module is graph.py:
from pathlib import Path
from import_deps import ModuleSet
def get_adjacency_list(path: str) -> dict[str, set[str]]:
pkg_paths = Path(path).glob('**/*.py')
module_set = ModuleSet([str(p) for p in pkg_paths])
d = {
m: module_set.mod_imports(m)
for m in sorted(module_set.by_name)
}
return d
Which gives output like this (details redacted):
{'foo.__init__': set(),
'foo.a': set(),
'foo.bar': {'foo.a',
'foo.d',
'foo.utils.j',
'foo.utils.k'},
'foo.c': {'foo.fancy_bar'},
'foo.d': set(),
'foo.e': {'foo.fancy_bar'},
'foo.f': {'foo.c'},
'foo.fancy_bar': {'foo.bar'},
'foo.g': {'foo.c'},
'foo.h': {'foo.c'},
'foo.i': {'foo.c'},
'foo.utils.__init__': set(),
'foo.utils.j': set(),
'foo.utils.k': set()}
foois the name of the packageutilsis a subpackage offoobaris an important module infoofancy_baris a module infoodoing something similar tobar- The module
ais a low-level component ofbar - The module
c,d,e,f,g,h, i` are core features
This was a code library I was working on (not a traditionally packaged Python package, but essentially treated as one).
When I drew out the diagram of the modules on a whiteboard,
it was clear to me that bar was provider to many of the other modules,
but of these fancy_bar stood out as it was in turn provider to c and e,
which meant that the core features (the single letter modules of foo)
were being provided by different modules, but all ultimately depended on bar.
In short, this stuck out to me when visualised,
and seemed to suggest that bar and fancy_bar should be refactored
in a more structured way:
- The features could go in a
featuresdirectory barandfancy_barcould go in abarsdirectory and both be accessed viabar
This is an example of where visualising the problem automatically (without manual whiteboarding) could lead me to see the suggested restructuring of the code faster.
Visualisation also made clear that:
- There are no 'orphaned' (standalone) modules
- The
amodule was indirectly imported into all of the features
Often a standalone node in the graph can indicate work to be done to connect a module up to the rest. It's also important to see the indirect components of modules, for example when wanting to test certain parts of the code in isolation (e.g. without touching deployed resources).
With just a little extra transforming we can get both in and out edges for each module:
def get_import_graph(path: str):
imports_in = get_adjacency_list(path)
imports_out = {}
for module, imports in imports_in.items():
for imported_name in imports:
imports_out.setdefault(imported_name, set())
imports_out[imported_name].add(module)
graph = {
module: {
"in": imps_in,
"out": imports_out.get(module, set()),
}
for module, imps_in in imports_in.items()
}
return graph
The full graph for the example above then looks like:
{'foo.__init__': {'in': set(), 'out': set()},
'foo.a': {'in': set(), 'out': {'foo.bar'}},
'foo.fancy_bar': {'in': {'foo.bar'},
'out': {'foo.e', 'foo.c'}},
'foo.c': {'in': {'foo.fancy_bar'}, 'out': set()},
'foo.bar': {'in': {'foo.a',
'foo.d',
'foo.utils.j',
'foo.utils.k'},
'out': {'foo.fancy_bar',
'foo.f',
'foo.g',
'foo.h',
'foo.i'}},
'foo.d': {'in': set(), 'out': {'foo.bar'}},
'foo.e': {'in': {'foo.fancy_bar'}, 'out': set()},
'foo.f': {'in': {'foo.bar'}, 'out': set()},
'foo.g': {'in': {'foo.bar'}, 'out': set()},
'foo.h': {'in': {'foo.bar'}, 'out': set()},
'foo.i': {'in': {'foo.bar'}, 'out': set()},
'foo.utils.__init__': {'in': set(), 'out': set()},
'foo.utils.j': {'in': set(), 'out': {'foo.bar'}},
'foo.utils.k': {'in': set(), 'out': {'foo.bar'}}}
We now have enough to draw the graph of module imports!
Network diagrams with graph-tool
I was delighted to find that an efficient successor to networkx/igraph has appeared in the past 2 or 3
years: graph-tool (available from conda-forge). It has a Graph class with a method
add_edge_list() which can take a list of pairs (i, j) which you can generate from an adjacency
list (reusing the get_adjacency_list function from earlier).
- Note that an adjacency list would not contain nodes with no edges
Before we can use that adjacency list, we need the nodes to be numeric, which I did by extracting all the names and numbering alphabetically from 0.
def numeric_adjacencies(adjacency_list: dict[str, set[str]]) -> dict[int, set[int]]:
# Collect all module names from keys and values of adjacency list
all_node_names = sorted({
*adjacency_list.keys(),
*[val for adj in adjacency_list.values() for val in adj]
})
node2int = {n: i for i, n in enumerate(all_node_names)}
return {
node2int[i]: set(node2int[j] for j in adj)
for i, adj in adjacency_list.items()
}
def make_directed_graph(path: str):
A = get_adjacency_list(path)
A_n = numeric_adjacencies(adjacency_list=A)
edges = [(j,i) for i in A_n for j in A_n[i]]
G = gt.Graph(directed=True)
G.add_edge_list(edges)
return G
This creates the graph, and to view it we call graph_draw (imported from graph_tool.draw).
Note the edges are reversed as (j,i) rather than (i,j) as the relation "importing from" is better
represented as an inward-bound edge rather than an outward one.
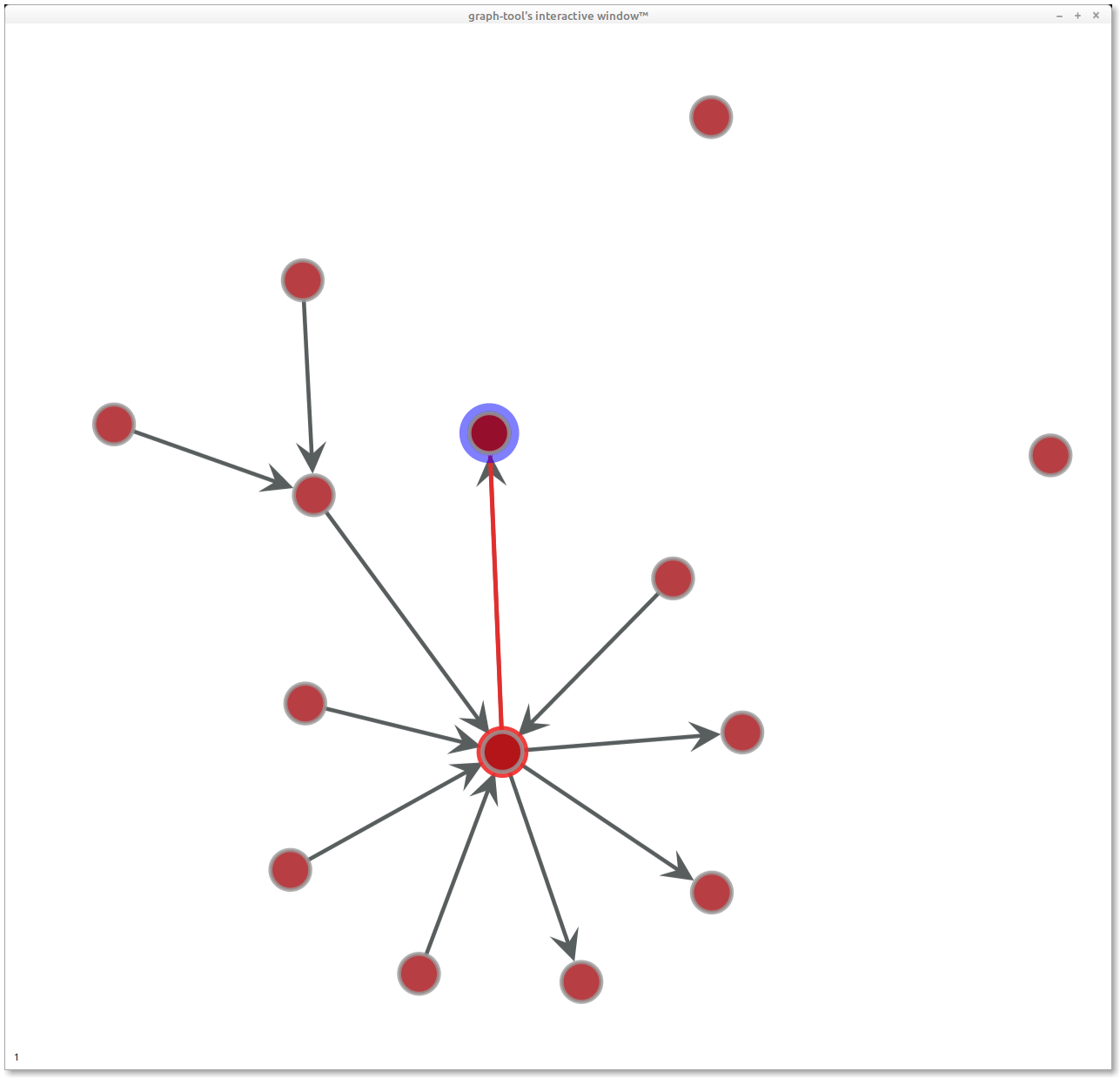
Looking good, but no labels yet.
To add labels, we can iterate over the vertices and add a vertex property "name", following the docs. At this point I refactored into a class and we're away.
dex
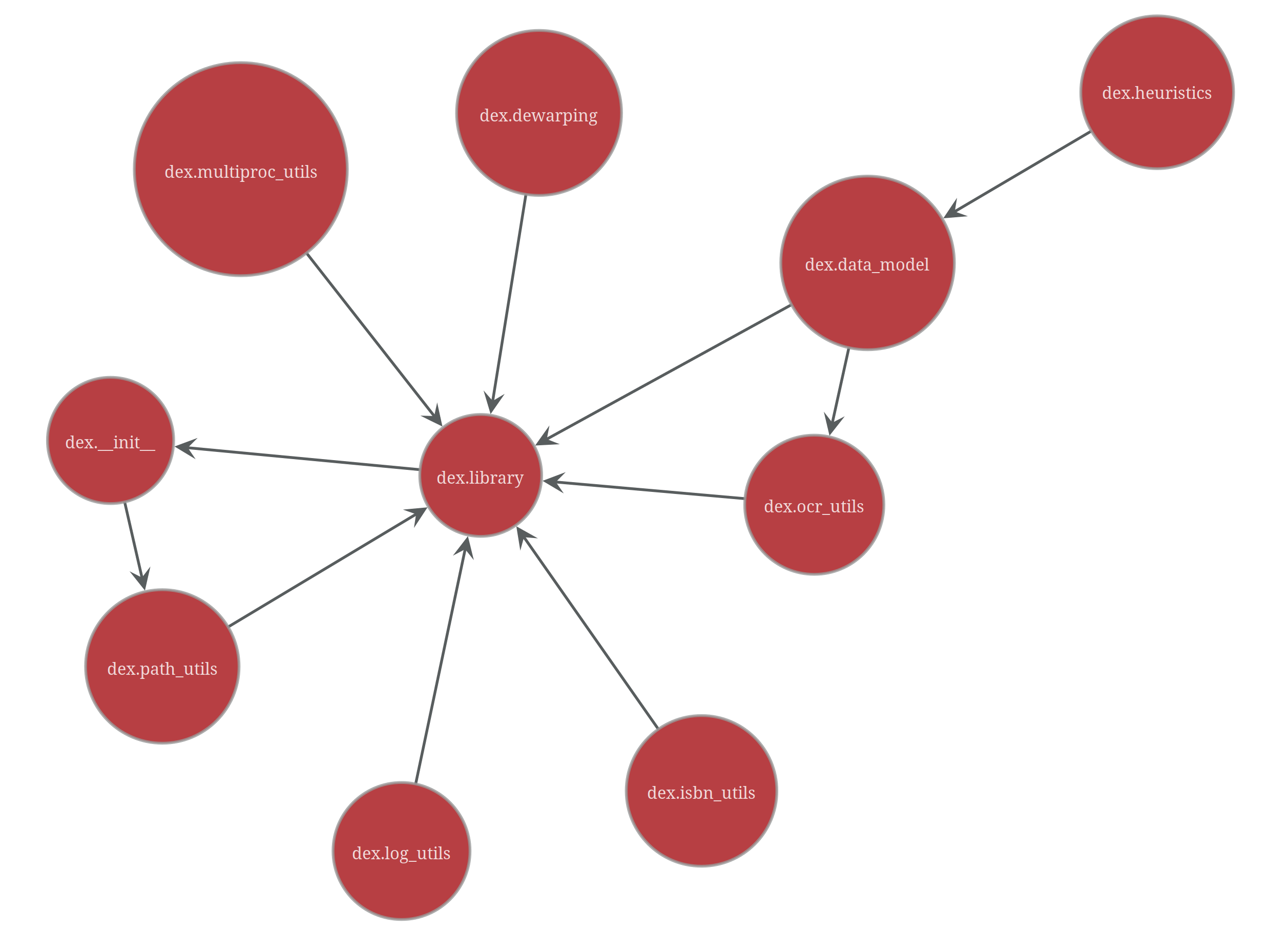
dex is a nice example, as the codebase remains fairly small (I stopped development on it after underwhelming results from OCR on book indexes).
On the left you can spot a circuit! The __init__ module imports the library into the package
namespace, while the path_utils module imports the package path from __init__, and lastly the
library module imports various paths built from this package path from path_utils.
This is perfectly fine from what I remember (does not cause a circular import error), as I recall
due to implicit package namespaces.
This view also suggests the logging utilities haven't gotten enough love outside of the library module.
range-streams
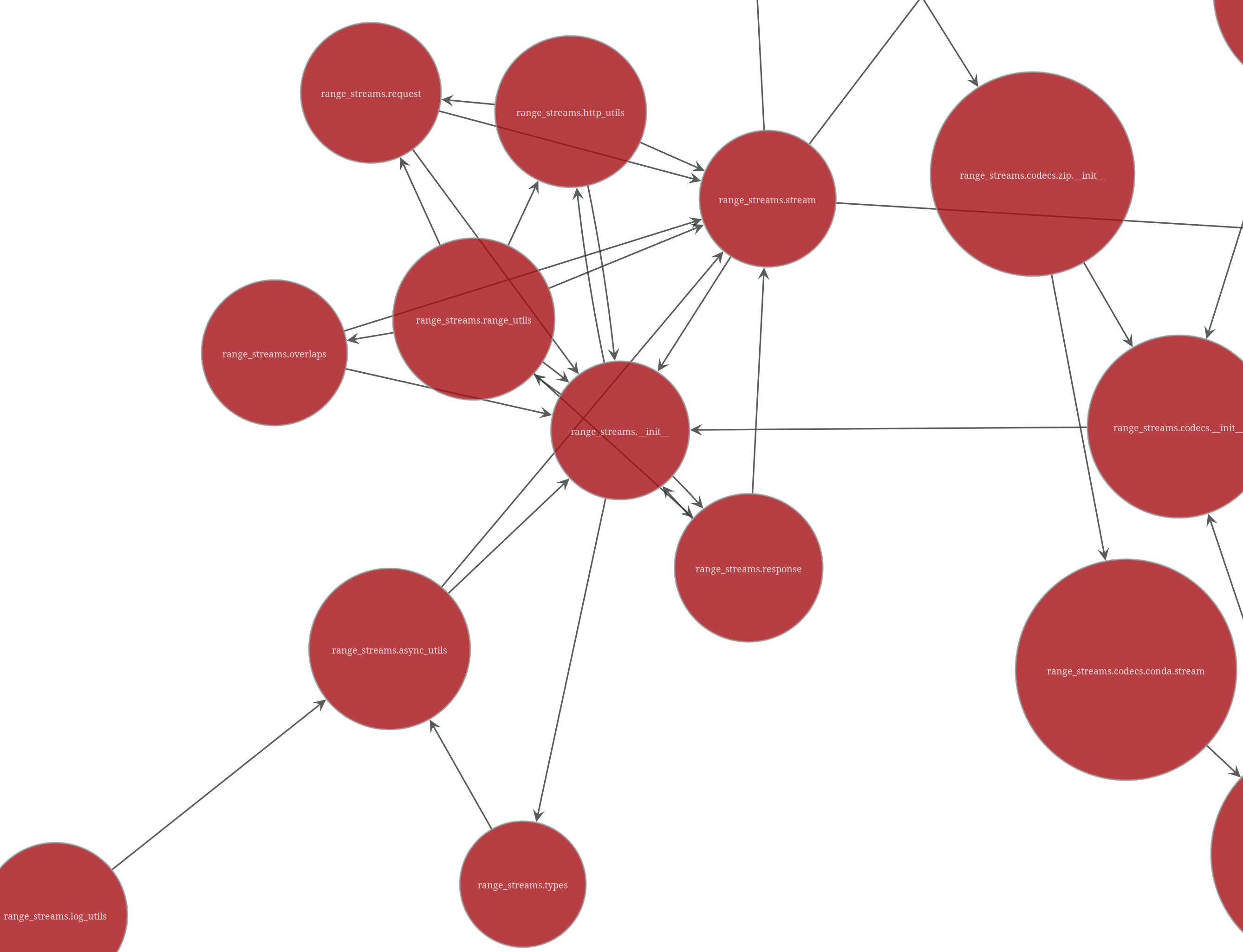
range-streams is a heavily documented library and it shows in the module imports, which look extremely busy on the network diagram.
The rest of the diagram (not shown) makes clear at a glance the purpose of the various parts,
e.g. the codecs.share subpackage is used to provide common utilities to both the zip and tar
codecs. There are 3 modules downstream of the stream module, etc. etc. This would all be very
useful for jogging memory or reviewing code someone else wrote and handed over to you to work on.
Other observations
Based on some code I'm not showing here, I also notice that __init__ modules
tend to be 'orphaned' nodes on the graph,
which indicates they're not particularly functional
(they are mainly used to specify an API for a library but this isn't necessary).
I'd like it if graph-tools would lay out the network with more 'direction',
so if there's one central node and some of the edges go in and some go out,
I'd prefer the layout to put the inward and outward nodes on different sides,
whereas it just intersperses them pretty much at random (maybe it's alphabetical).
I'd also like the edges to show what gets imported, if that wouldn't crowd the graph too much.
To prevent the nodes becoming too large, I abbreviated the package name so that even if your input
had full qualnames (foo.bar) the network displayed shows them as relative module names
(.bar). Spaces and newlines didn't make the text wrap.
(TBC!)
This post is the 2nd of Mapping inter-module imports with importopoi, a series covering how I developed a library to depict the graph of module imports for any Python package (or set of scripts) for clarity about how package computation is structured at a glance, and subsequent ease of jumping in to edit or review a particular part. Read on for discussion of TDD for software that runs on code
